Research Project: DNA Binding Domains
Computational and biophysical investigation into nuclear receptor and p53 DNA binding interfaces for targeted drug development.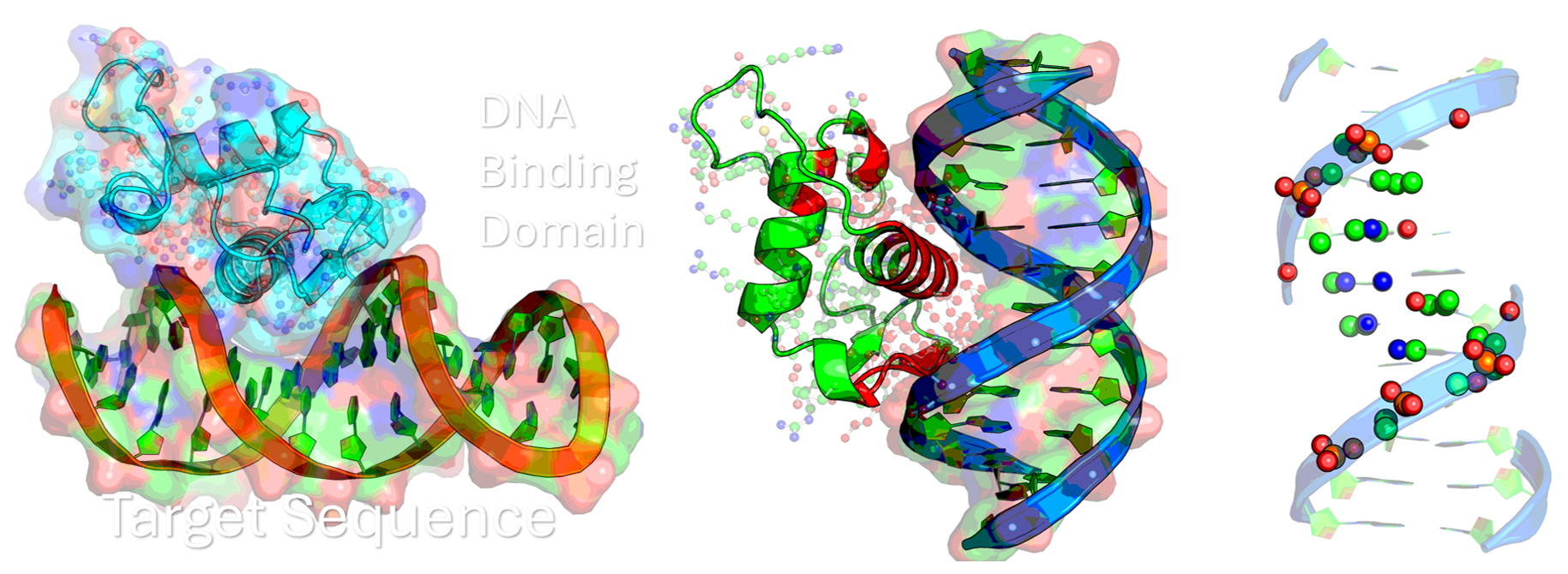
Computational and biophysical investigation into nuclear receptor and p53 DNA binding interfaces for targeted drug development.
Investigating the structural dynamics of proline-rich domains (PRDs) in key proteins like p53 and Tau using molecular dynamics simulations to understand disease mechanisms.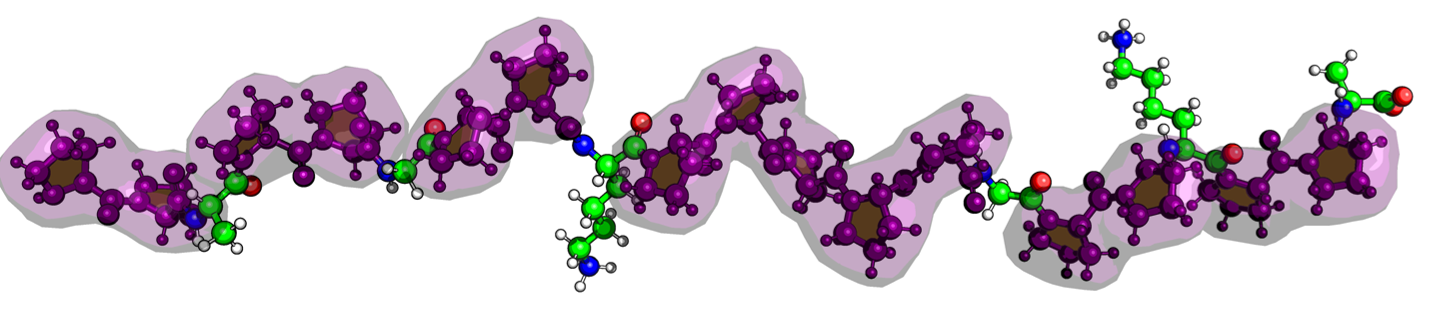
Utilizing virtual screening, molecular dynamics, and free energy calculations to discover and optimize ligands for orphan nuclear receptors (e.g., NR2F family) with the goal of developing first-in-class targeted therapies.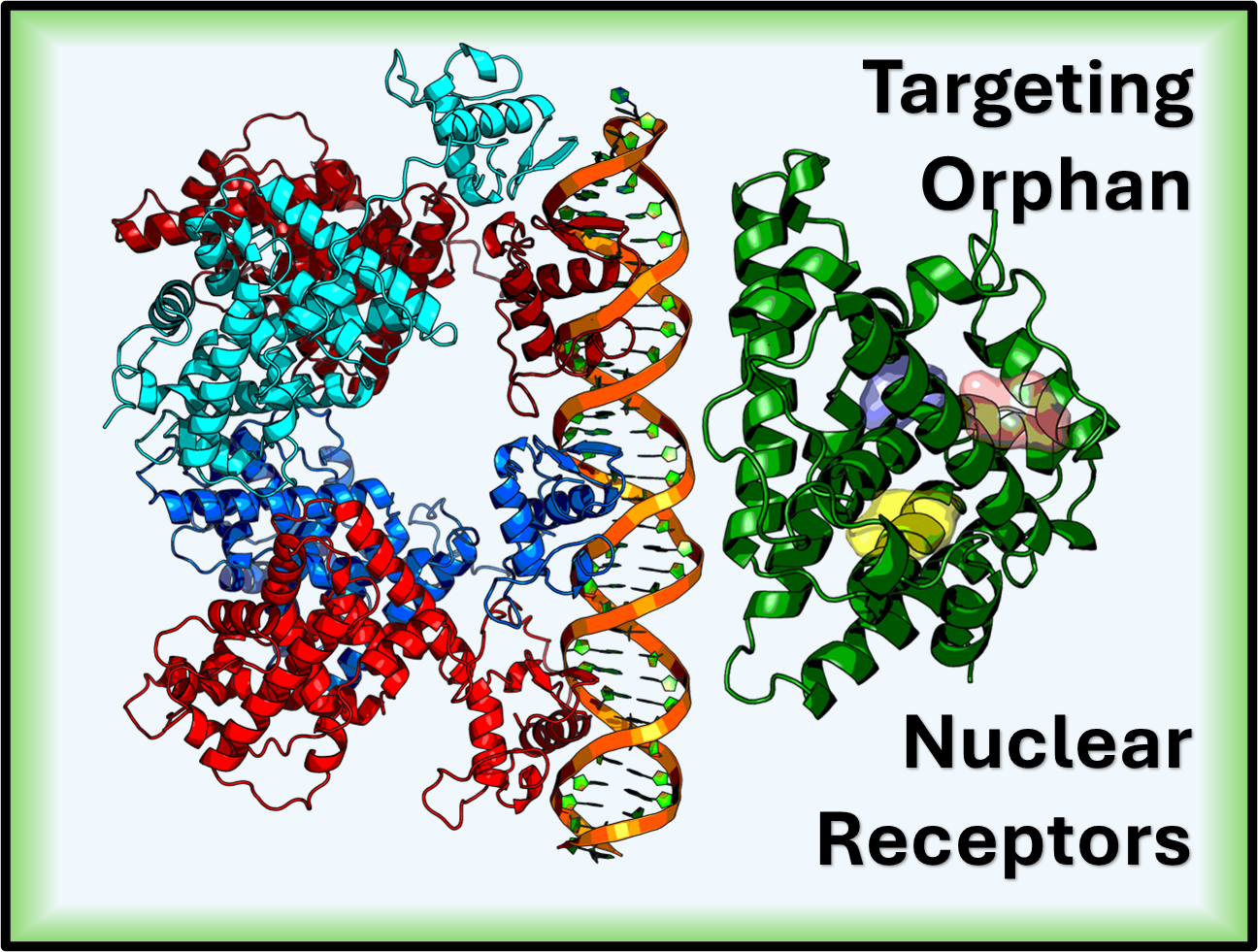
Developing advanced computational protocols that integrate Dimensionality Reduction and Clustering to create highly representative conformational ensembles for accurate NMR chemical shift predictions in Intrinsically Disordered Proteins.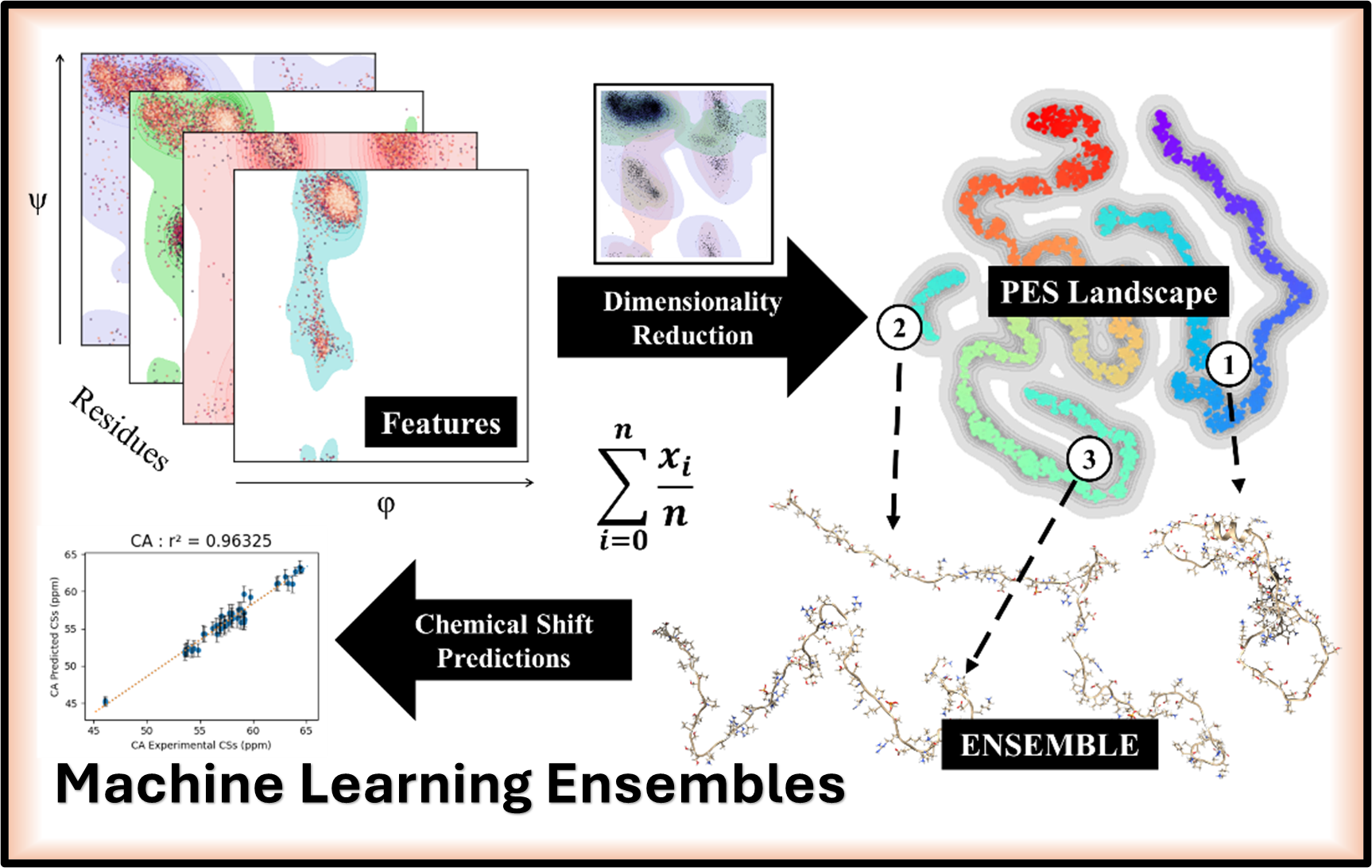
Developing and refining advanced computational schemes (MD/ADMA/DFT/QM/MM) to achieve high-accuracy NMR chemical shift predictions for Intrinsically Disordered Proteins (IDPs) and to deconstruct the 31P NMR signal in modified nucleic acids.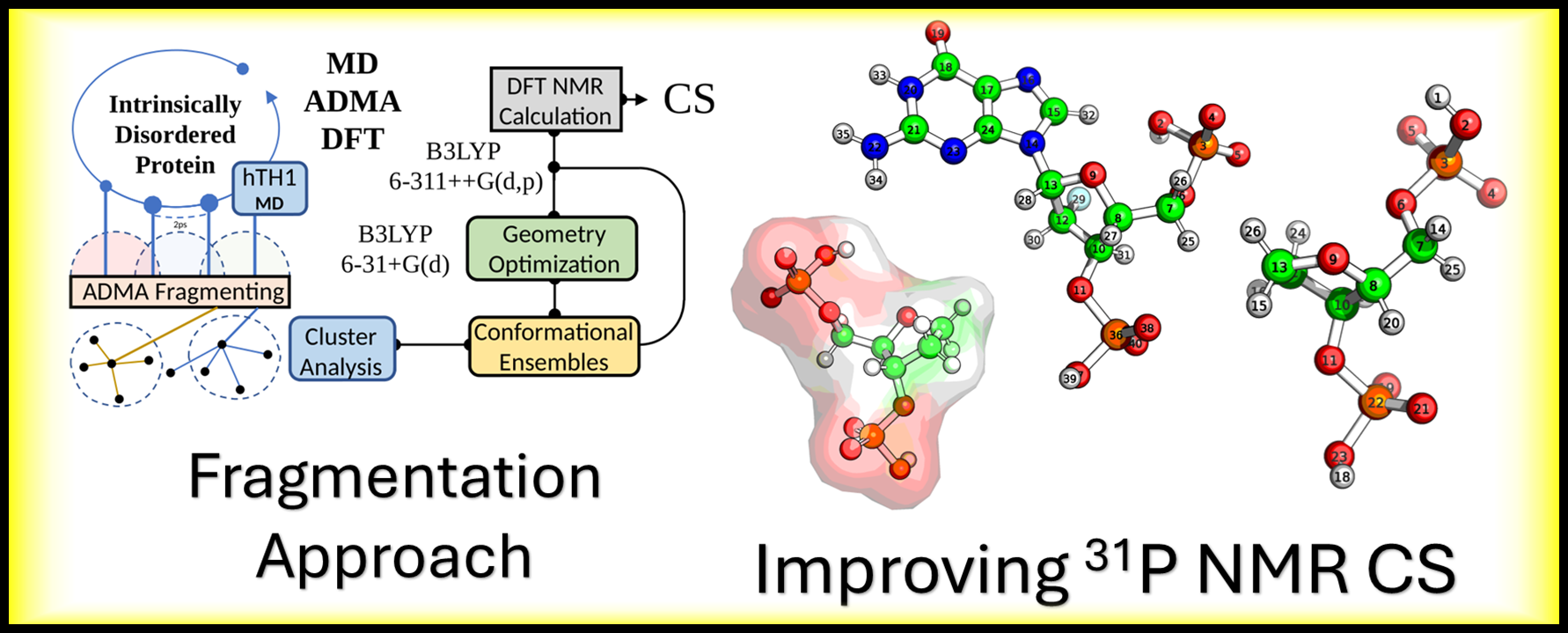
Investigating how phosphorylation and phosphomimetic modifications influence protein structure and dynamics, focusing on biomolecular size, secondary structure changes, and effects on protein-protein interactions. 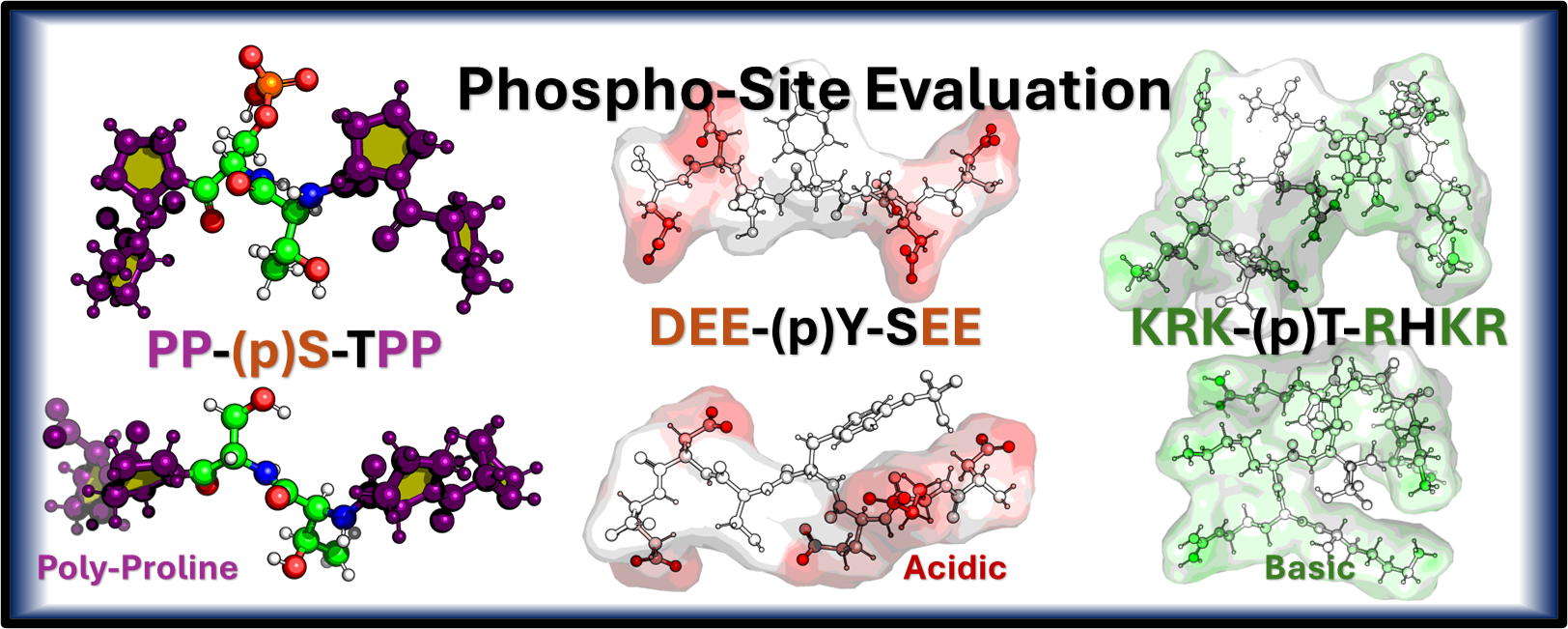
A comprehensive molecular dynamics investigation across all four p53 IDRs (TAD1, PRD, PTL, REG) to elucidate how local sequence features, post-translational modifications, and domain context govern the protein’s conformational landscape, allostery, and critical regulatory functions.
Published in Concepts in Magnetic Resonance, 2019
This paper discusses a method for creating 3D printed models of NMR data using Mathematica and a 3D printer, which can serve as a helpful teaching tool for students to understand complex information in NMR spectra.
Download here
Published in Physical Chemistry and Chemical Physics, 2022
This paper is about the generation of NMR chemical shifts ab-inito from IDP trajectories using a multiscale approach.
Download here
Published in Energies, 2024
The long-term unsustainability of petroleum-based products is addressed by investigating the direct molecular dynamics simulation of the pyrolysis of methyl linoleate (a biodiesel substitute) to understand the atomistic details of its decomposition and identify alternative, renewable pathways to produce valuable petrochemical precursors like ethene and propene.
Download here
Published in Journal of Chemical Information and Modeling, 2024
This study streamlines the sampling procedure for predicting NMR chemical shifts (CS) in intrinsically disordered proteins (IDPs) by merging dimensionality reduction (DR) (specifically using solvent-accessible surface area (SASA)) and clustering algorithms (CA) to generate clustered ensembles (CEs) that outperform traditional sequential ensembles.
Download here
Published in Journal of Chemical Information and Modeling, 2024
This study introduces an approach to better understand the conformational ensemble (CoE) of intrinsically disordered proteins (IDPs) by decomposing the ensemble of the model peptide histatin 5 into groups separated by their radius of gyration (Rg) using a combination of small-angle X-ray scattering (SAXS) and all-atomistic molecular dynamics simulations.
Download here
Published in Journal of Chemical Theory and Computation, 2024
This study uses fully atomistic explicit water molecular dynamics simulations to explore the functional landscape of the intrinsically disordered p53 regulatory domain, revealing a detailed understanding of how varying the number of phosphorylated amino acids impacts its conformational dynamics, flexibility, structural effects, protein interactivity, and energy landscapes.
Download here
Published in Journal of Chemical Theory and Computation, 2024
This paper is about an investigation made into the disordered regions of p53 using molecular dynamics and machine learning techniques.
Download here
Published in Archiv der Pharmazie (2025), 2025
An integrated computational approach led to the discovery of Hit-3 (CSC089939231), a new nonsteroidal Takeda G protein-coupled receptor 5 (TGR5) modulator, with molecular dynamics simulations suggesting its activation is supported by a hydrogen bond interaction with Tyr240.
Download here
Published in Biophysical Journal (2025), 2025
All-atomistic molecular dynamics simulations of the Tau(210-240)/BIN1 SH3 complex reveal that additional phosphate charges reduce salt-bridge formation by up to 32% and cause a strong preference in Tau for contact with the distal and n-Src loops instead of the RT-Src loop.
Download here
Published:
The p53 protein is also known as the ”Guardian of the Genome” as it can trigger cell cycle arrest, DNA repair, and apoptosis. Mutations in p53 is quite common in the development of cancer. The protein is also quite complex with multiple domains, and also contains four signficantly disorderd regions as seen below.
University Sponsored Tutoring, Missouri State University, Department of Chemistry, 2016
Worked with students through collaboration from the faculty (Professor Siebert) to provide supplemental support for the Organic Chemistry course.
Lab Instructor, Missouri State University, Department of Chemistry, 2018
Instructed students on hands-on wet chemistry related to their lessons in Organic Chemistry I and II.
Practical Classes, Charles University, Faculty of Pharmacy, 2025
Taught practical classes focused on utilizing simple publicly available office tools to overcome minor obstacles in computational work and data presentations.
Teaching Assistant, Charles University, Faculty of Pharmacy, Department of Inorganic and Organic Chemistry, 2025
Provided supplementary instruction and support for students enrolled in the introductory Calculus and Statistics courses.