Research Project: DNA Binding Domains
Computational and biophysical investigation into nuclear receptor and p53 DNA binding interfaces for targeted drug development.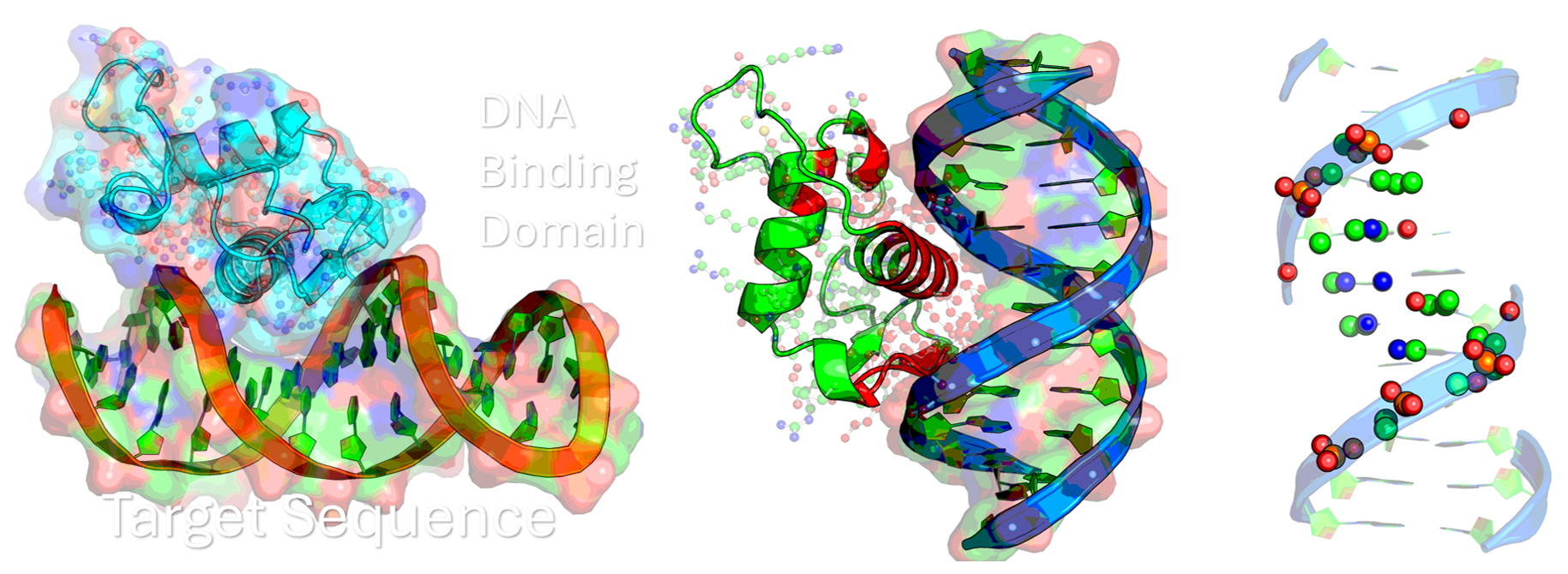
Computational and biophysical investigation into nuclear receptor and p53 DNA binding interfaces for targeted drug development.
Investigating the structural dynamics of proline-rich domains (PRDs) in key proteins like p53 and Tau using molecular dynamics simulations to understand disease mechanisms.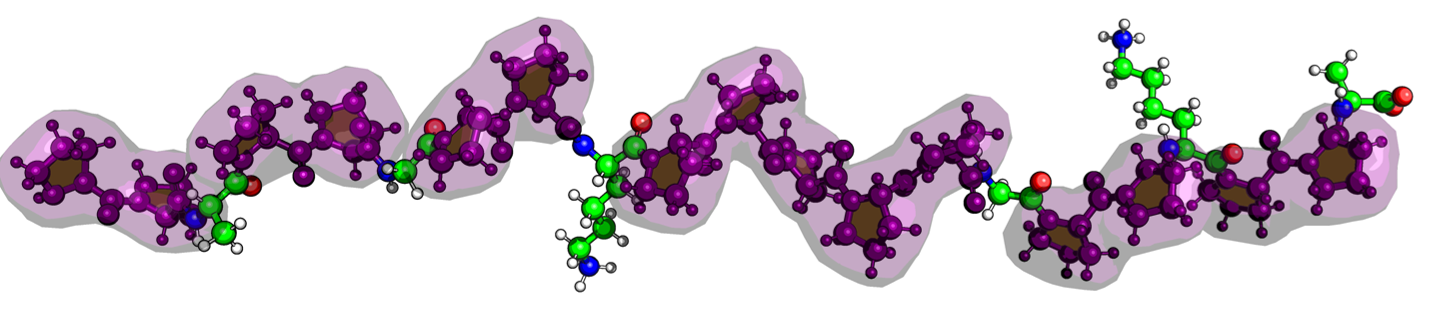
Utilizing virtual screening, molecular dynamics, and free energy calculations to discover and optimize ligands for orphan nuclear receptors (e.g., NR2F family) with the goal of developing first-in-class targeted therapies.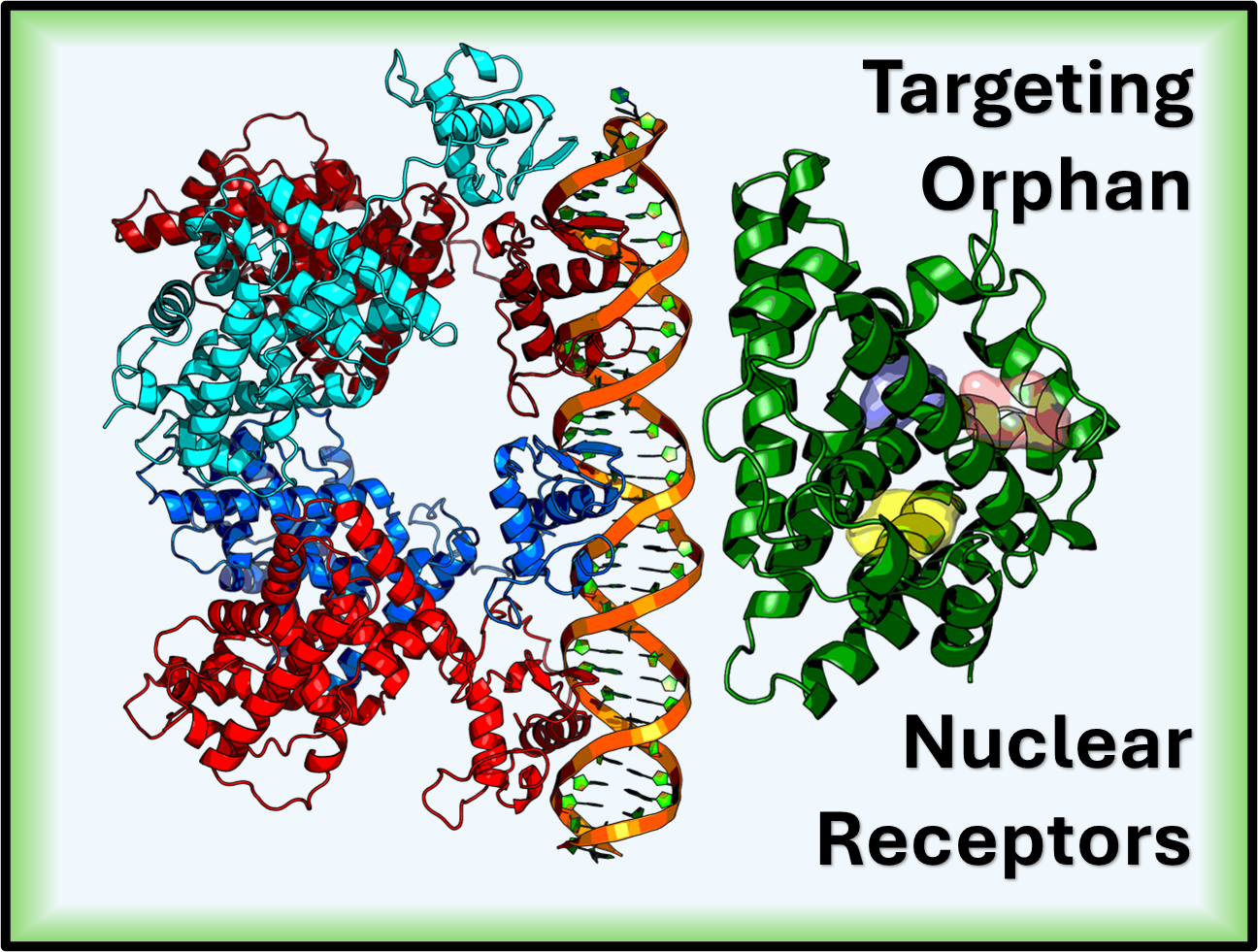
Developing advanced computational protocols that integrate Dimensionality Reduction and Clustering to create highly representative conformational ensembles for accurate NMR chemical shift predictions in Intrinsically Disordered Proteins.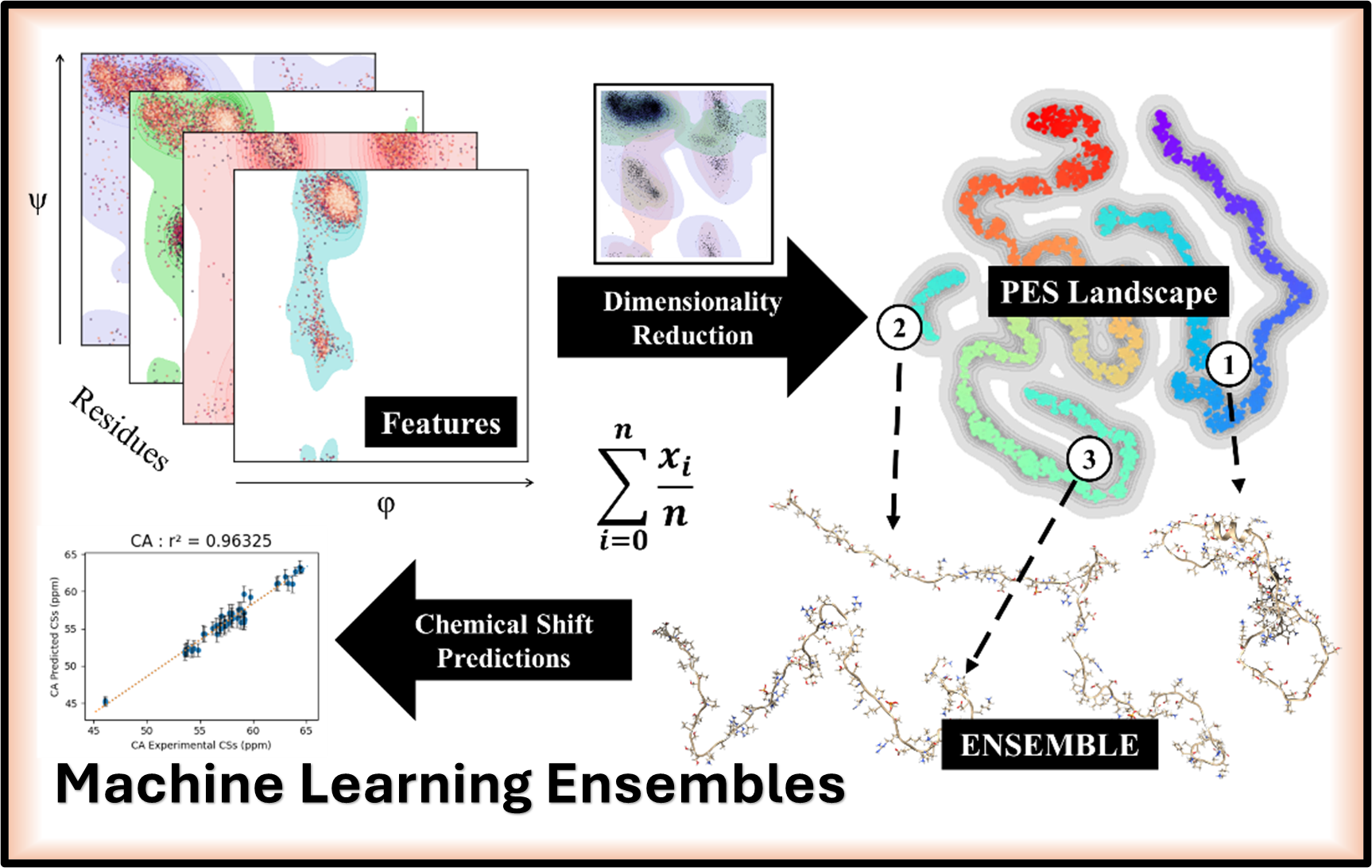
Developing and refining advanced computational schemes (MD/ADMA/DFT/QM/MM) to achieve high-accuracy NMR chemical shift predictions for Intrinsically Disordered Proteins (IDPs) and to deconstruct the 31P NMR signal in modified nucleic acids.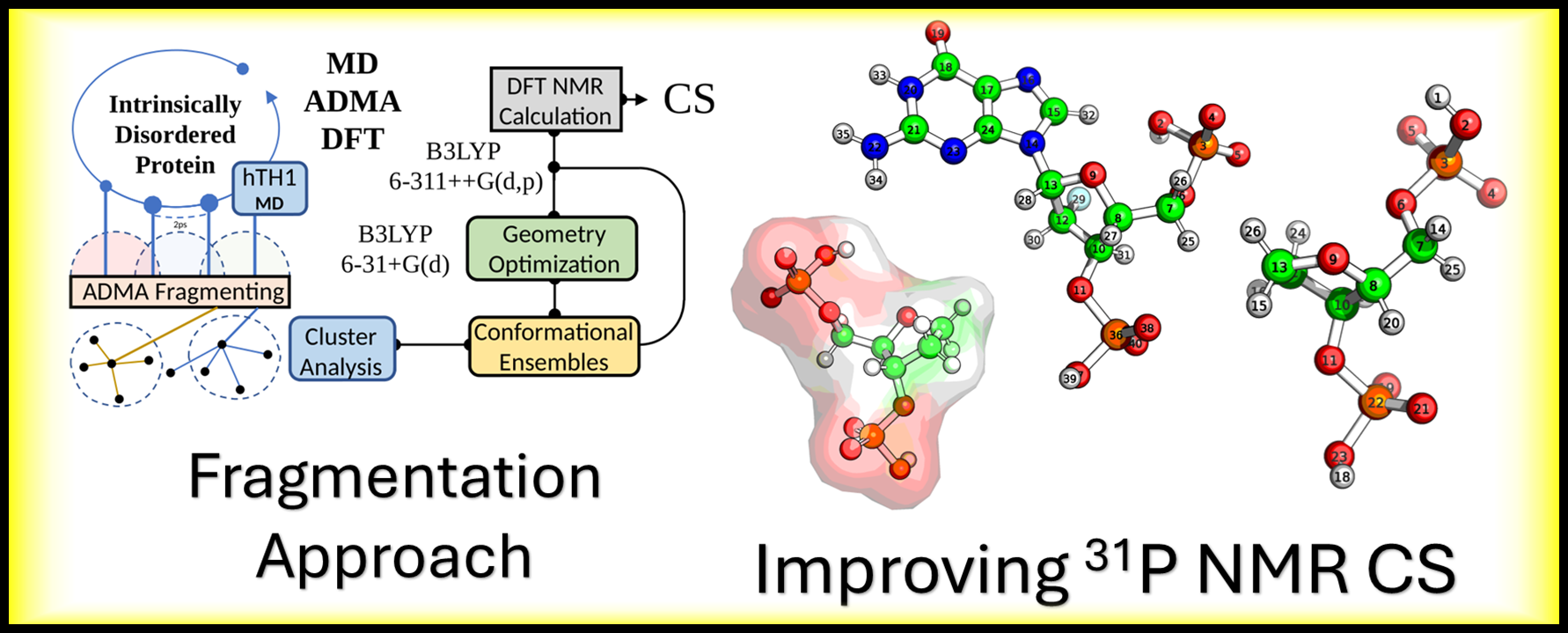
Investigating how phosphorylation and phosphomimetic modifications influence protein structure and dynamics, focusing on biomolecular size, secondary structure changes, and effects on protein-protein interactions. 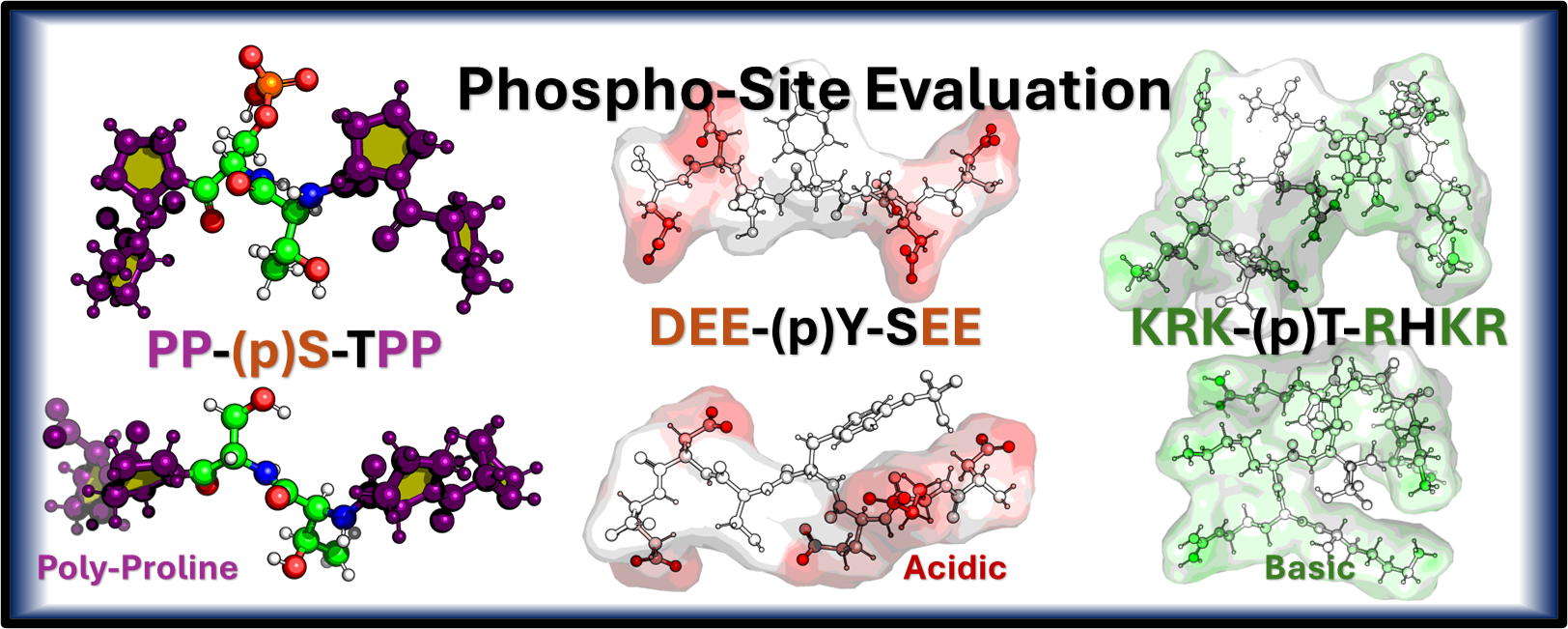
A comprehensive molecular dynamics investigation across all four p53 IDRs (TAD1, PRD, PTL, REG) to elucidate how local sequence features, post-translational modifications, and domain context govern the protein’s conformational landscape, allostery, and critical regulatory functions.