Research Project: DNA Binding Domains
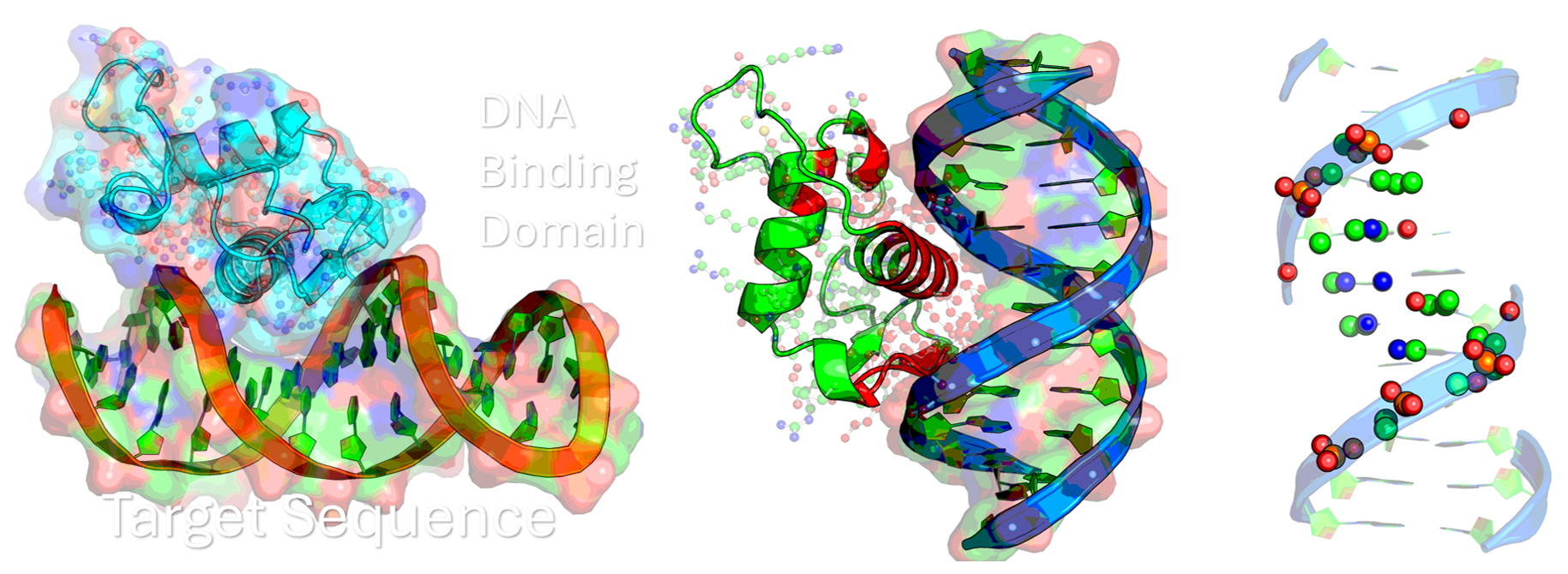
Project Goal: Investigating DNA Binding Domains for Targeted Therapeutics
This research project aims to computationally and structurally investigate the DNA-binding domains (DBDs) of critical nuclear receptors and transcription factors.
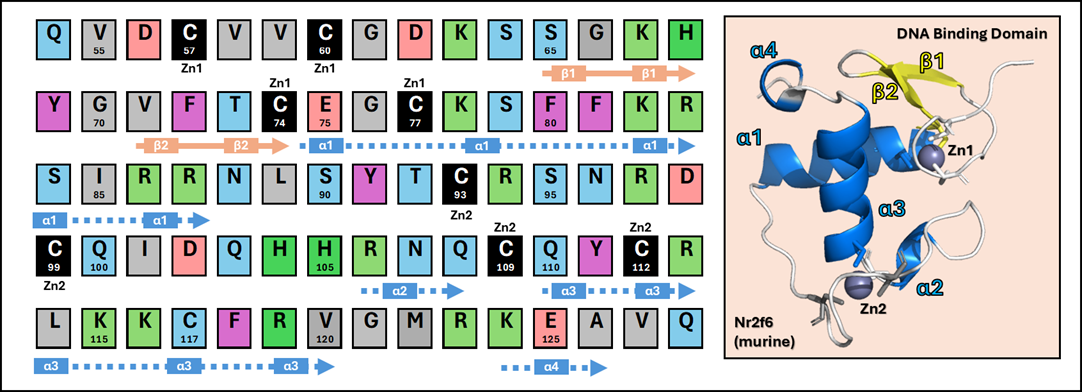
Step 1 - Investigate the Interface: Characterize the molecular interface between DNA and specific nuclear receptors, such as the NR2F family (e.g., NR2F1, NR2F2), or the tumor suppressor p53. This involves understanding the precise residues and structural motifs responsible for sequence-specific binding and regulation.
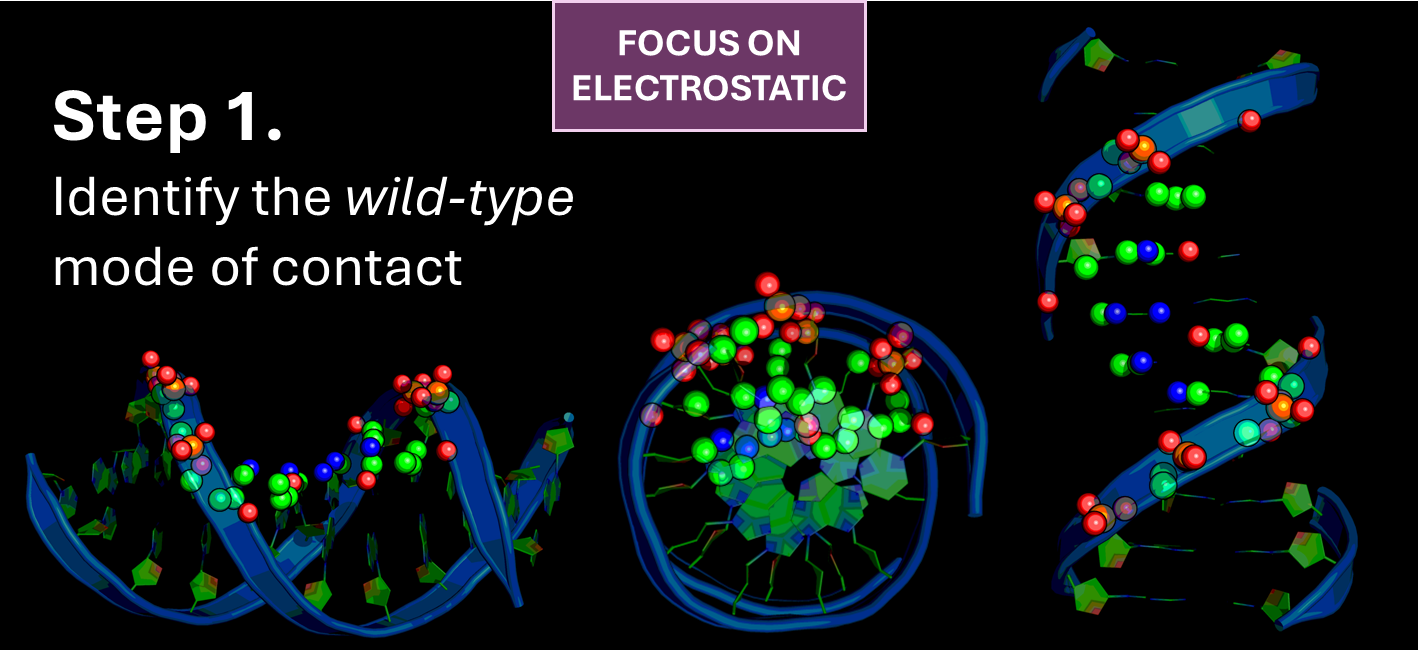
Step 2 - Identify Targetable Cavities: Utilize advanced simulation and docking techniques to identify and characterize cryptic or allosteric binding cavities within these protein-DNA complexes.
Step 3 - Targeted Drug Development: Develop targeted drug development strategies focused on identifying or designing small molecules that can fit into these identified cavities to modulate (enhance or disrupt) the receptor’s DNA binding activity. This is a crucial step towards creating novel therapeutics for diseases driven by nuclear receptor dysfunction or p53 mutation. Building upon the structural insights from Steps 1 and 2, this phase focuses on rational drug design aimed at modulating the DNA-binding behavior of nuclear receptors and transcription factors. The goal is to develop small molecules that selectively interact with cryptic or allosteric cavities identified in the protein-DNA interface, thereby influencing transcriptional regulation in a therapeutically beneficial manner.
Strategic Objectives:
- Allosteric Modulation: Design ligands that bind to non-canonical pockets to induce conformational shifts, altering the receptor’s affinity or specificity for DNA.
- Disruption of Pathogenic Interactions: In cases like mutant p53, develop inhibitors that prevent aberrant DNA binding or restore wild-type function.
- Enhancement of Receptor Activity: For underactive receptors (e.g., NR2F1 in neurodevelopmental disorders), identify activators that stabilize the DNA-bound conformation.
Implications of this Research
This project holds significant therapeutic promise by moving beyond traditional ligand-binding domains to target the regulatory mechanisms at the protein-DNA interface . By discovering and targeting allosteric pockets on DNA-binding domains, we create a new avenue for developing first-in-class small molecules capable of selectively fixing gene expression programs. This approach could lead to novel treatments for cancers (by restoring p53 function), metabolic disorders, and immune diseases (by modulating nuclear receptor activity) where genetic transcription is dysregulated.
Project Collaborators

Professor Petr Pavek at Charles University's Faculty of Pharmacy specializes in Nuclear Receptor (NR) pharmacology. His lab provides essential experimental validation for computational predictions on DNA-binding domains (DBDs). Using cell-based systems like reporter gene assays and qPCR, his team confirms how our simulated compounds modulate the transcriptional activity and DNA binding specificity of targeted nuclear receptors or the p53 protein, ensuring functional relevance for drug development.
