Targeting Orphan Nuclear Receptors for Novel Therapies
Project Goal: Discovery of Ligands for Orphan Nuclear Receptors (ONRs)
This project is dedicated to accelerating targeted drug development by computationally characterizing and identifying novel ligands for challenging Orphan Nuclear Receptors (ONRs), such as members of the NR2F family. These receptors are transcription factors whose endogenous ligands and precise regulatory mechanisms remain unknown, making them powerful yet untapped therapeutic targets for metabolic disorders, cancer, and developmental diseases.
| Receptor Name | Gene Symbol | Receptor Family | Proposed Function |
|---|---|---|---|
| COUP-TFII | NR2F2 | NR2F | Development, angiogenesis, metabolism |
| DAX-1 | NR0B1 | NR0B | Reproductive axis regulation |
| SHP | NR0B2 | NR0B | Bile acid metabolism, transcriptional repression |
| NURR1 | NR4A2 | NR4A | Dopaminergic neuron development |
| NOR-1 | NR4A3 | NR4A | Cell proliferation, apoptosis |
| GCNF | NR6A1 | NR6A | Germ cell development, embryogenesis |
Computational Strategy
We employ a multi-stage, high-resolution computational workflow to conquer the challenges presented by these structurally enigmatic receptors:
- Structural Dynamic Characterization (MD Simulations):
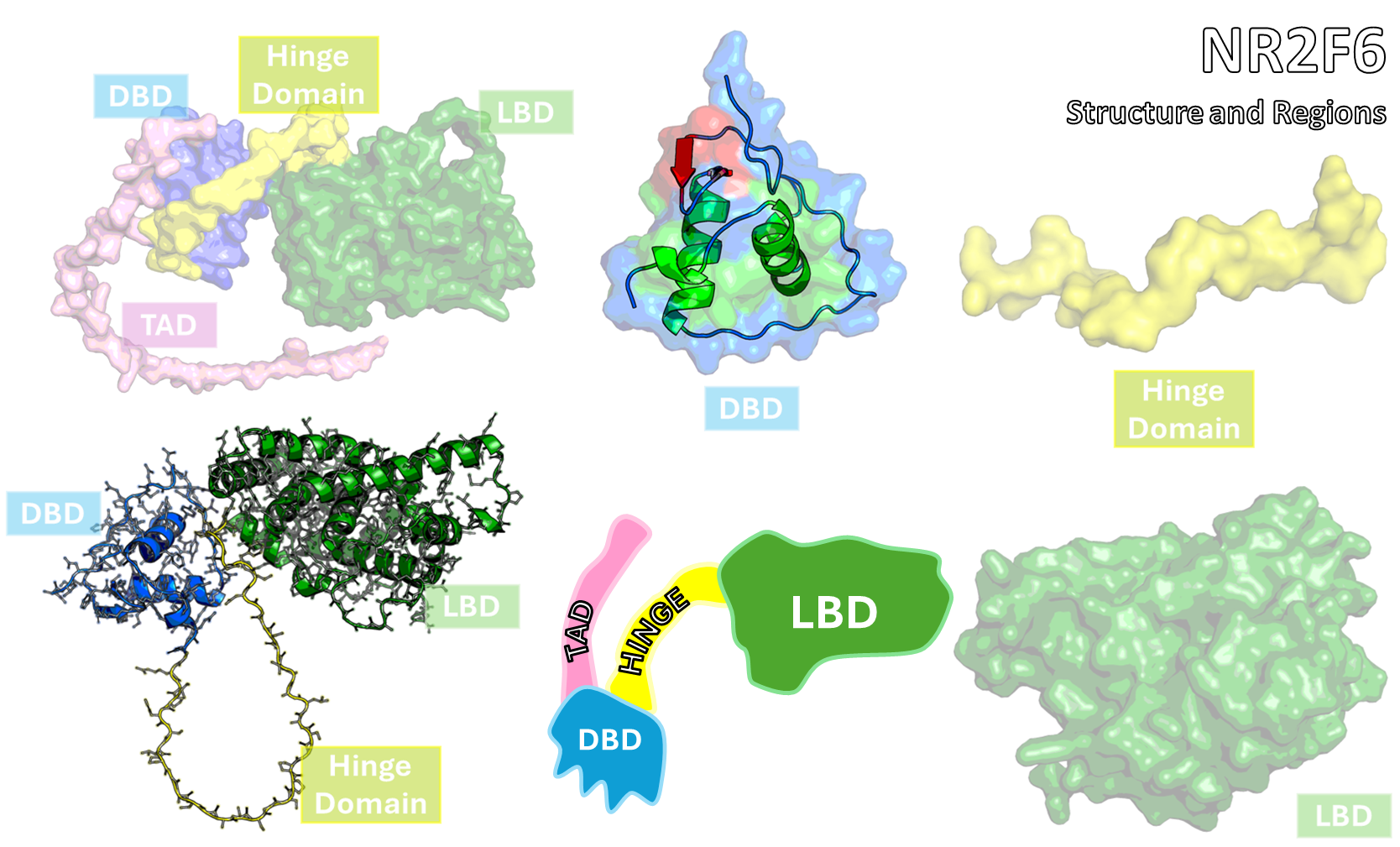
- Since ONRs often exhibit high flexibility or require binding to DNA (as explored in Project 1) to stabilize their ligand-binding domains (LBDs), traditional static crystal structures are insufficient.
- We use extensive Molecular Dynamics (MD) simulations to sample the complete conformational ensemble of the target receptor, identifying both known and cryptic allosteric pockets relevant for small-molecule binding.
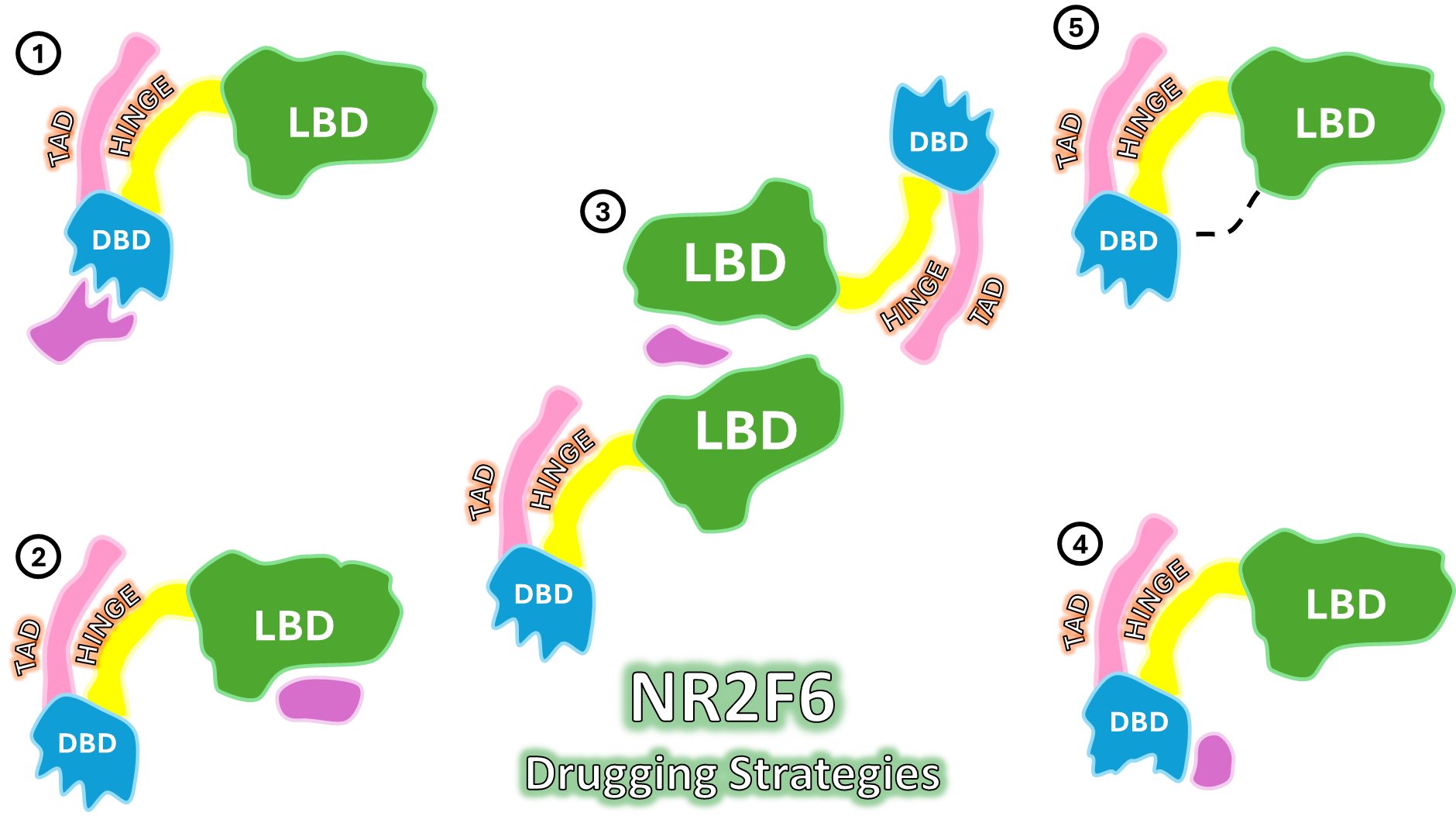
- Advanced Virtual Screening and Docking:
- We utilize the refined conformational ensemble to perform Ensemble Docking against vast small-molecule libraries. This maximizes the probability of finding initial hit compounds that can accommodate the receptor’s intrinsic flexibility.
- This approach focuses on identifying molecules that bind with high affinity and selectively modulate the receptor’s transcriptional activity.
- Ligand Optimization via Free Energy Calculations:
- Initial hits are rigorously prioritized using advanced techniques like MMGBSA/MM-PBSA and Free Energy Perturbation (FEP) calculations.
- This computational gold standard provides highly accurate predictions of ligand binding affinities ($\Delta G$) and pinpoints critical atomic-level interactions, guiding the iterative design and synthesis of highly potent lead compounds.
Therapeutic Impact
Success in this project provides a mechanism to de-orphanize these critical receptors, creating therapeutic pipelines aimed at diseases where conventional targets have proven ineffective. By precisely modulating the activity of the NR2F family or other ONRs, we open pathways to first-in-class drugs that target the root transcriptional drivers of complex human pathology.
Project Collaborators

Prof. Petr Pavek at Charles University is a critical collaborator whose lab focuses on the pharmacology of Nuclear Receptors (NRs) involved in drug metabolism and cancer. Using assays like reporter gene assays and qPCR, his data confirms the affinity and activity of identified ligands for Orphan Nuclear Receptors (ONRs). This crucial biological feedback guides our computational efforts to refine virtual hits into potent therapeutic leads.
