Prolines Have Never Had So Much Style
Published:
I’ve been deep in the world of Intrinsically Disordered Proteins (IDPs). These aren’t the neat, rigid structures you see in textbooks; they’re dynamic, flexible, and utterly central to life, handling critical tasks like cellular signaling and regulating protein-protein interactions.
Proline is one of the most structurally unique amino acids, and its rigid, five-membered pyrrolidine ring is a major player in protein folding, structure, and function. Yet, visualizing this ring clearly amidst the complex 3D structure of a protein can be surprisingly tricky.
The Importance of Visualizing the Pyrrollidine Ring
The core structure of a proline, specifically the pyrrollidine ring, governs its behavior. A proline can exist in either a cis or trans isomeric state around the peptide bond, a switch that can completely change a protein’s function. This image is to show the importance of understanding the structure of the proline.
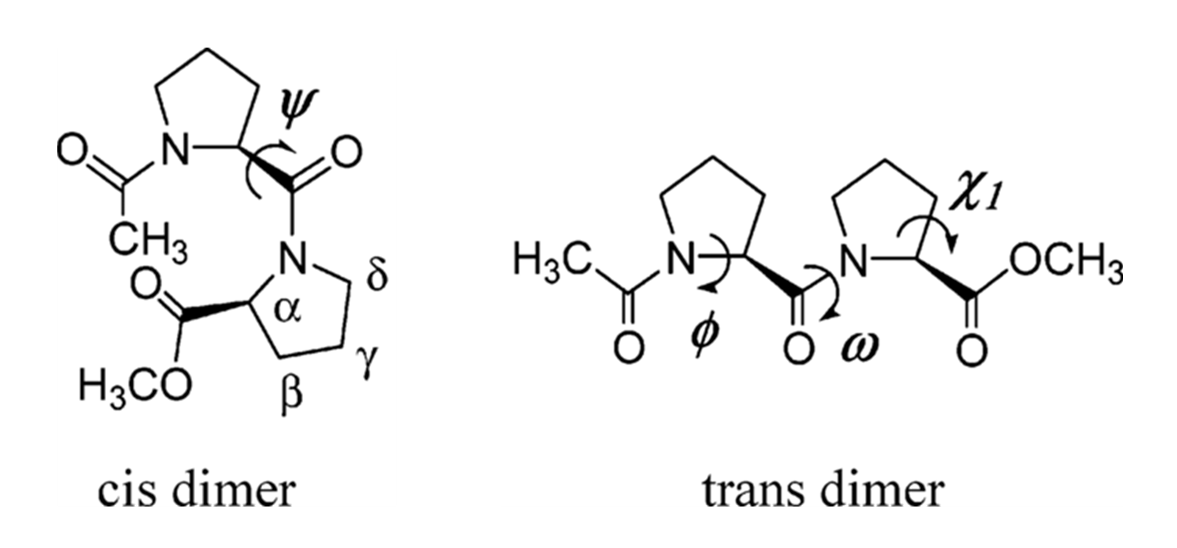
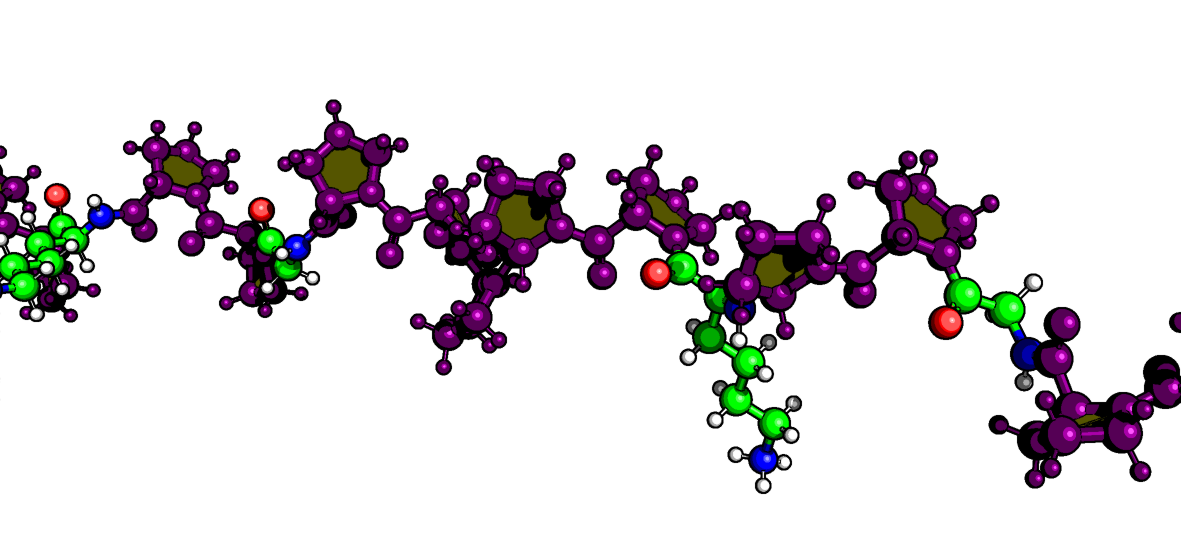
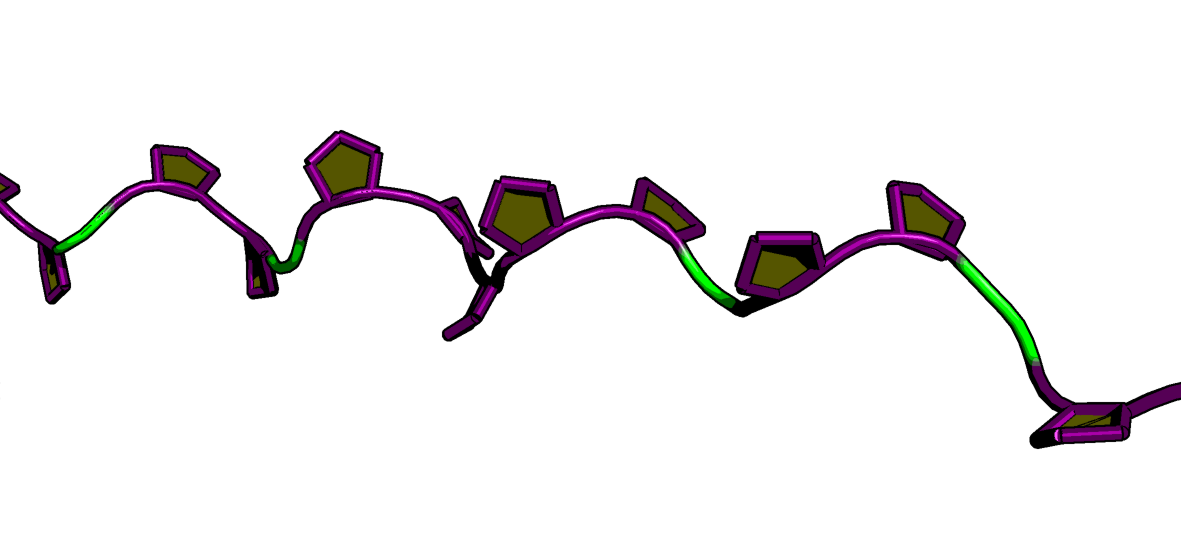
When you look at a protein structure, the proline ring (composed of the N, $\text{C}\alpha$, $\text{C}\beta$, $\text{C}\gamma$, and $\text{C}\delta$ atoms) is often just a regular part of the stick model. It can be hard to distinguish it from the backbone or side chains of other residues. To truly study its conformation—like its famous puckering or its role in cis/trans isomerism—you need a visualization that highlights this critical ring in a dedicated, high-impact way.
To solve this visualization problem, I wrote a Python script. This PyMOL extension uses Custom Graphic Objects (CGOs) to give every proline ring the spotlight it deserves.
Hover over the image to see the transformation
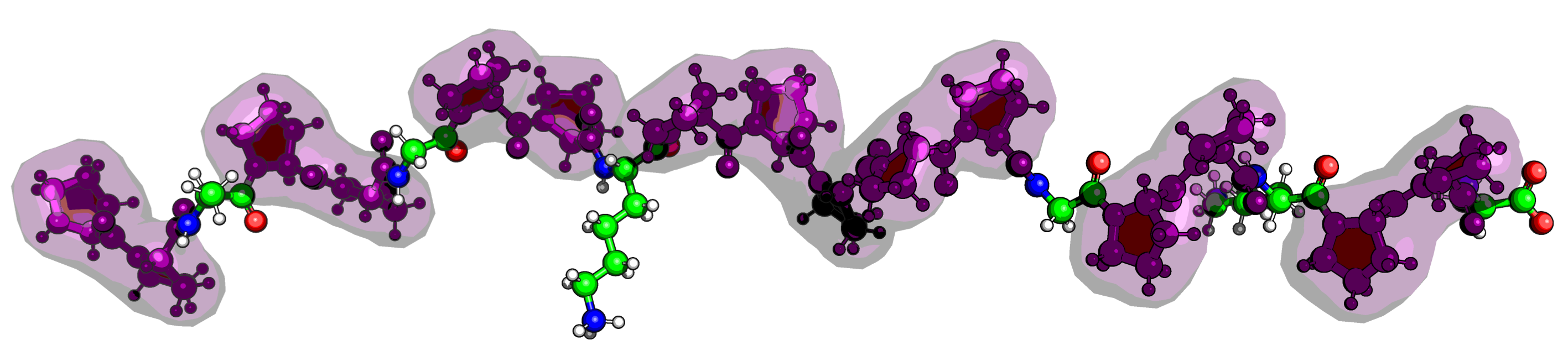
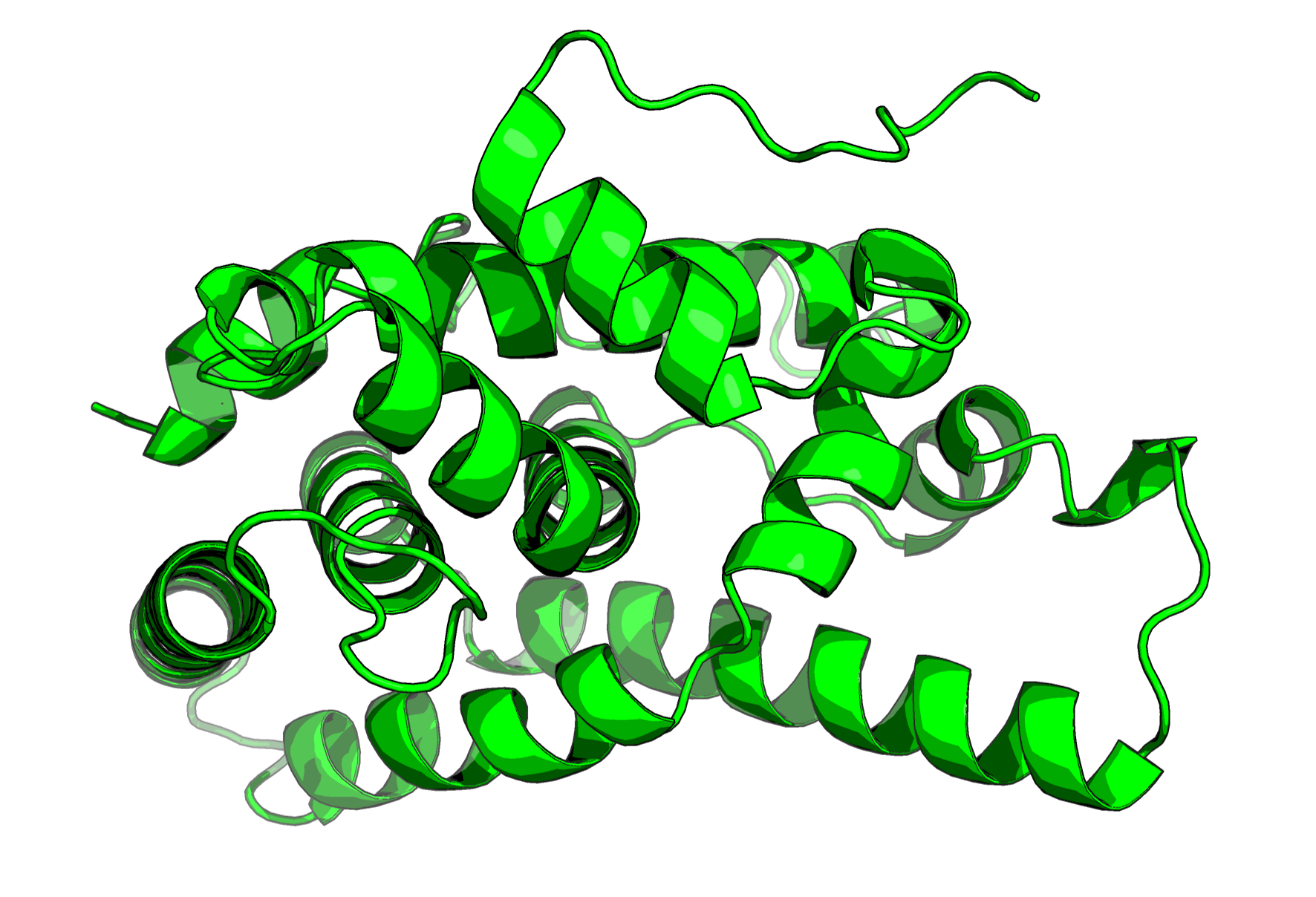
Here’s a breakdown of what the script does:
1. High-Contrast Ring Highlighting
The heart of the script is the creation of CGOs that target the five ring atoms ($\text{N}$, $\text{C}\alpha$, $\text{C}\beta$, $\text{C}\gamma$, $\text{C}\delta$):
Yellow Filled Pentagon: A solid, yellow-filled CGO pentagon is drawn across the face of the ring. This instantly defines the ring’s plane and makes its structural conformation easy to spot.
Purple Tube Outline: Thick purple CGO cylinders are drawn along the edges of the ring. This high-contrast outline ensures the feature pops against the protein structure.
2. Full Residue Styling
Beyond the ring itself, the script colors the entire proline residue purple and adjusts the display mode for clarity:
- It hides the default cartoon to minimize clutter.
- It automatically adds hydrogens and displays both hydrogen and non-hydrogen atoms as size-adjusted spheres, giving you full atomic detail.
- It creates an optional, semi-transparent purple surface (
prings) over the ring atoms to show solvent accessibility.
To use the script, you must first load it into PyMOL. Once loaded, run the function with your object name:
Next, run the script:
# Assuming you named your script fancyProlines.py
run fancyProlines.py
# Run the function on your structure
proline_pentagons("peptide")
Note: Remember to replace “peptide” with the name of the object you want to apply it to.
The script also works for full structured proteins, instantly highlighting all prolines in a complex fold.
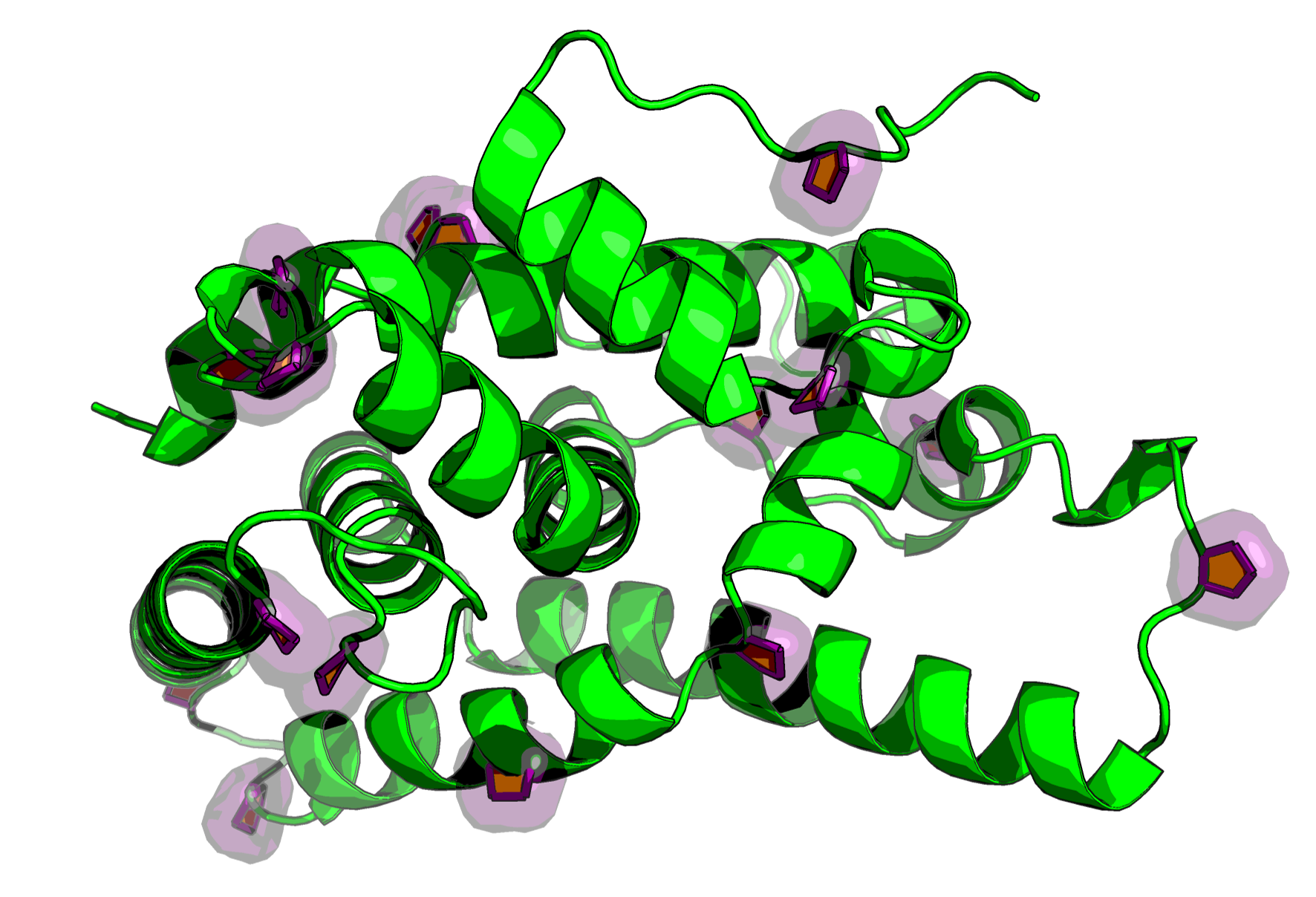
Copy and paste the script below into a python script of your naming in a location you can access, and run the script like above.
Full Python Script:
import pymol
from pymol import cmd, cgo
import numpy as np
def calculate_normal(P0, P1, P2):
V01 = np.array(P1) - np.array(P0)
V02 = np.array(P2) - np.array(P0)
normal = np.cross(V01, V02)
norm = np.linalg.norm(normal)
if norm == 0:
return [0.0, 0.0, 0.0]
return (normal / norm).tolist()
def proline_pentagons(selection="all", cgo_name="proline_rings_cgo", color=(1.0, 1.0, 0.0), tube_radius=0.2, filled=True):
cmd.set("two_sided_lighting", 0)
cmd.set("ray_trace_mode", 3)
cmd.set("ray_trace_color", "black")
cmd.set("stick_radius", 0.15, selection)
cmd.hide("cartoon", selection)
cmd.h_add(selection)
cmd.show("sticks", selection)
cmd.show("spheres", selection)
cmd.set("sphere_scale", 0.2, "hydrogens")
cmd.set("sphere_scale", 0.3, "not hydrogens")
yellow = (1.0, 1.0, 0.0)
purple = (0.5, 0.0, 0.5)
tube_color = purple
tube_name = "proline_tubes_cgo"
ring_selection_name = "prings"
cmd.set_color("proline_purple", purple)
cmd.color("proline_purple", f"({selection}) and resn PRO")
prolines = cmd.get_model(f"({selection}) and resn PRO", 1)
if not prolines.atom:
return
unique_prolines = set()
for atom in prolines.atom:
identifier = (atom.segi, atom.chain, atom.resi)
unique_prolines.add(identifier)
if not unique_prolines:
return
full_cgo = []
tube_cgo = []
ring_selection_strings = []
if not filled:
full_cgo.extend([cgo.LINEWIDTH, tube_radius * 10, cgo.COLOR, color[0], color[1], color[2]])
ring_atoms = ['N', 'CA', 'CB', 'CG', 'CD']
for segi, chain, resi in unique_prolines:
base_sel = f"(segi '{segi}' and chain '{chain}' and resi {resi} and resn PRO)"
coords = {}
missing_atom = False
atom_names = []
for name in ring_atoms:
sel_str = f"{base_sel} and name {name}"
try:
xyz = cmd.get_atom_coords(sel_str, state=1)
if xyz is None:
missing_atom = True
break
coords[name] = xyz
atom_names.append(name)
except:
missing_atom = True
break
if missing_atom:
continue
num_atoms = len(atom_names)
if num_atoms != 5:
continue
ring_selection_strings.append(f"{base_sel} and name N+CA+CB+CG+CD")
if filled:
full_cgo.extend([cgo.COLOR, yellow[0], yellow[1], yellow[2]])
P0 = coords[atom_names[0]]
P1 = coords[atom_names[1]]
P2 = coords[atom_names[2]]
P3 = coords[atom_names[3]]
P4 = coords[atom_names[4]]
normal = calculate_normal(P0, P1, P2)
full_cgo.extend([
cgo.NORMAL, normal[0], normal[1], normal[2],
cgo.BEGIN, cgo.TRIANGLE_FAN,
cgo.VERTEX, P0[0], P0[1], P0[2],
cgo.VERTEX, P1[0], P1[1], P1[2],
cgo.VERTEX, P2[0], P2[1], P2[2],
cgo.VERTEX, P3[0], P3[1], P3[2],
cgo.VERTEX, P4[0], P4[1], P4[2],
cgo.END
])
else:
full_cgo.extend([cgo.BEGIN, cgo.LINES])
for i in range(num_atoms):
start_name = atom_names[i]
end_name = atom_names[(i + 1) % num_atoms]
start_coord = coords[start_name]
end_coord = coords[end_name]
full_cgo.extend([
cgo.VERTEX, start_coord[0], start_coord[1], start_coord[2],
cgo.VERTEX, end_coord[0], end_coord[1], end_coord[2],
])
full_cgo.extend([cgo.END])
for i in range(num_atoms):
start_name = atom_names[i]
end_name = atom_names[(i + 1) % num_atoms]
start_coord = coords[start_name]
end_coord = coords[end_name]
tube_cgo.extend([
cgo.CYLINDER,
start_coord[0], start_coord[1], start_coord[2],
end_coord[0], end_coord[1], end_coord[2],
tube_radius,
tube_color[0], tube_color[1], tube_color[2],
tube_color[0], tube_color[1], tube_color[2]
])
if full_cgo:
cmd.delete(cgo_name)
cmd.load_cgo(full_cgo, cgo_name)
cmd.enable(cgo_name)
if tube_cgo:
cmd.delete(tube_name)
cmd.load_cgo(tube_cgo, tube_name)
cmd.enable(tube_name)
if ring_selection_strings:
combined_selection = " or ".join(ring_selection_strings)
cmd.delete(ring_selection_name)
cmd.create(ring_selection_name, combined_selection)
cmd.hide("spheres", ring_selection_name)
cmd.hide("sticks", ring_selection_name)
cmd.hide("cartoon", ring_selection_name)
cmd.show("surface", ring_selection_name)
cmd.set_color("prings_color", purple)
cmd.color("prings_color", ring_selection_name)
cmd.set("transparency", 0.5, ring_selection_name)
cmd.extend("proline_pentagons", proline_pentagons)